Build the Semantic Layer (Entity Embeddings + Metadata Linking)
Introduction
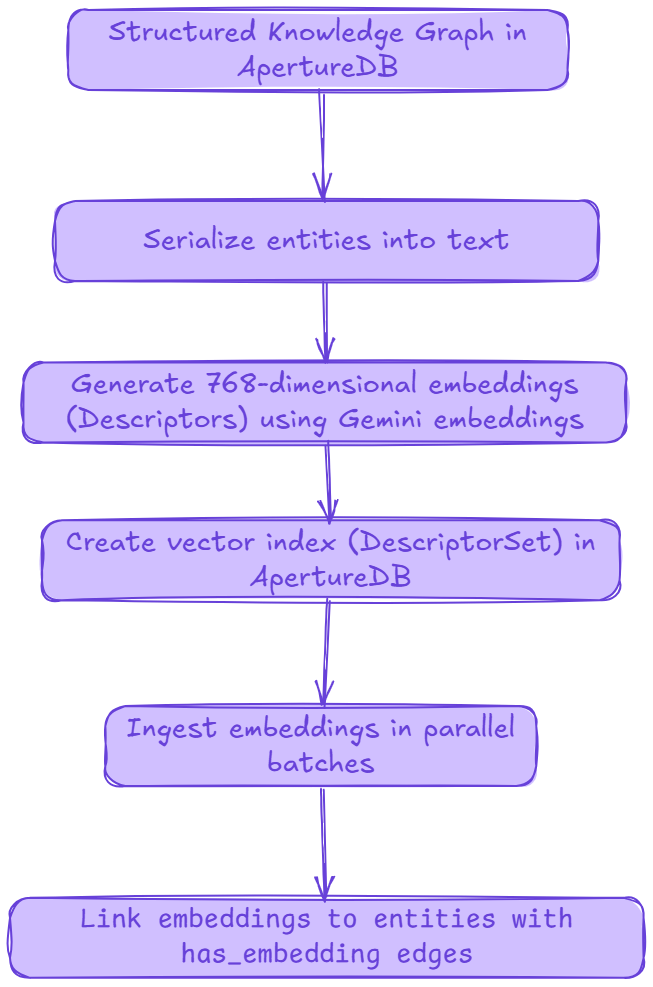
In the previous parts (Part 1 and Part 2) of this series we used Google’s Gemini to auto-extract class schemas, concrete entities, and their properties from PDF source material, then batch-ingested everything into an ApertureDB instance. That gave us a fully structured symbolic knowledge graph. We then added explicit relationships and visualized the graph. Now we’re ready to turn that graph into a hybrid retrieval substrate by layering semantic vector embeddings directly onto the entities you already stored. This is the critical first step toward GraphRAG: dense vector similarity lets us find semantically related entities; the graph lets us traverse contextually rich neighborhoods around them. Together, they dramatically improve recall, grounding, and explanation in downstream RAG workloads.
ApertureDB is purpose-built for multimodal AI/graph workloads: it stores structured metadata (your entities + relationships) alongside blobs, embeddings, and indexes optimized for similarity search; exactly what we need to bind vectors to graph nodes and query across both signals. (ApertureDB Documentation)
In this segment we’ll do three things:
- Generate embeddings (Descriptors) for every entity in our existing graph using Gemini embeddings and package them with minimal metadata.
- Create an ApertureDB vector index (DescriptorSet) sized to the embedding dimensionality.
- Ingest + link: load embeddings at scale using ApertureDB’s ParallelLoader, store source entity metadata on each descriptor, and create has_embedding connections so we can hop from a vector hit back into the graph.
Notebook & Data: As before, code below is trimmed for readability. The full, executable Colab notebook (with setup cells, error handling, logging, etc.) and the complete Github repo are linked. The repo also contains some sample data you can use to run the notebooks. You will find that there are two versions of the notebook:
- The cloud version involves signing up on ApertureDB Cloud and configuring your instance there.
- The local udocker version involves setting up the DB instance locally using udocker (which allows running Docker containers in constrained environments like Colab notebooks) - no sign-up needed.
A Note on Scale & Inline Pipelines
In many real‑world projects you would likely embed while you are building the graph - ApertureDB’s loaders make that straightforward by letting you stream raw entities, blobs, and their embeddings in the same parallel ingest job, so a million‑node graph with tens of millions of vectors can be built end‑to‑end without a second pass (see docs).
But the opposite scenario is just as common: you might inherit an already‑constructed graph (or decide to experiment with a new embedding model) and only later realize you want to add or refresh vector data. Here too, the ParallelLoader uses exactly the same API: simply add an AddDescriptor entry for every entity row and run the ingest again.
In this tutorial series we decouple the steps on purpose: first to keep each post focused, and second to highlight ApertureDB’s native support for both graph and vector workloads. Whether you embed inline during graph creation or revisit the graph to try different models, the mechanics remain identical.
What You Should Already Have (Prerequisites)
You should be starting from an ApertureDB instance populated with:
- Entity classes + properties auto-derived from your document(s).
- Deduplicated entity instances, each with a unique id property.
- (Optional but recommended) The original PDF stored as a Blob.
- Relationships between entities.
If you haven’t completed those steps, pause here and work through Knowledge Graph Automation Part 1 and Part 2 - they automate nearly everything: upload a PDF, extract structure via Gemini, clean it, and batch-ingest into ApertureDB.
GraphRAG Phase 1 Overview: From Entities → Semantic Embeddings
Here’s the high-level flow we’ll implement in this post:
- Fetch entities from ApertureDB (all classes, all properties).
- Synthesize a text “document” per entity by concatenating its name, class, and property key-values. These become embedding inputs.
- Batch embed the documents using Google’s gemini-embedding-001 model (we’ll use 768-dim vectors for a balance of quality vs. storage/search cost).
- Create an ApertureDB DescriptorSet (vector index), ingest embeddings at scale, and create has_embedding edges back to source entities so semantic hits are graph-navigable.
Environment Setup
Install required packages (ApertureDB client, Google GenAI SDK, utilities) and pull credentials from your Colab secrets store (or set up udocker and Google Drive if you’re not using ApertureDB Cloud).
%pip install -q aperturedb google-genai
import os, json, time
import numpy as np
from typing import Any, Dict, List
from google.colab import userdata # or your own secrets loader
from aperturedb.CommonLibrary import create_connector
from aperturedb.ParallelLoader import ParallelLoader # used later
from google import genai
from google.genai import types
# Credentials (stored in Colab "secrets" / userdata)
google_api_key = userdata.get("GOOGLE_API_KEY")
# Connect to ApertureDB
client = create_connector(key=db_key)
Preparing Entity Data for Embedding
Why synthesize entity documents? Embedding models expect text. Your graph entities are structured key-value rows; we need a reliable textual serialization that captures discriminative properties (name, class, key attributes) so semantically similar entities land near each other in vector space. Including the class label often helps cluster related concepts and improves downstream filtering/re-ranking.
Fetch all entities from ApertureDB
We first read the schema to discover valid (non-internal) classes, then issue a FindEntity query per class requesting all properties. Internal classes (like _Blob) are skipped to avoid errors and noise.
def fetch_entities(client):
"""Fetch all user-defined entities (skip internal classes)."""
# 1. Get schema
schema_query = [{"GetSchema": {}}]
schema_response, _ = client.query(schema_query)
if (not schema_response
or "entities" not in schema_response[0]["GetSchema"]):
print("Error: Could not retrieve schema.");
return []
all_class_names = schema_response[0]["GetSchema"]["entities"]["classes"].keys()
valid_class_names = [c for c in all_class_names if not c.startswith("_")]
all_entities = []
for class_name in valid_class_names:
q = [{
"FindEntity": {
"with_class": class_name,
"results": {"all_properties": True}
}
}]
resp, _ = client.query(q)
ents = resp[0].get("FindEntity", {}).get("entities", [])
for e in ents:
e["class"] = class_name # convenience
all_entities.extend(ents)
print(f"Fetched {len(all_entities)} total entities.")
return all_entities
Serialize entities into embedding documents
We build a minimal but information-rich string: Entity: <name>. Class: <class>. key1: val1. key2: val2. You can enrich/curate which fields to include; keep it consistent so embeddings are comparable. We currently include all available entity properties that had been extracted by the LLM in knowledge graph creation.
def create_entity_documents(entities):
"""Return [{'entity_id', 'class', 'document'}, ...]"""
docs = []
for e in entities:
parts = [
f"Entity: {e.get('name', '')}.",
f"Class: {e.get('class', 'N/A')}."
]
for k, v in e.items():
if k in ("_uniqueid", "name", "class"):
continue
if v is not None:
parts.append(f"{k}: {v}.")
docs.append({
"entity_id": e.get("id"),
"class": e.get("class"),
"document": " ".join(parts)
})
print(f"Created {len(docs)} documents for embedding.")
return docs
Run it.
all_entities = fetch_entities(client)
docs_to_embed = create_entity_documents(all_entities) if all_entities else []
print(docs_to_embed[0])
Sample (truncated):
{
"entity_id": 208,
"class": "Application Architecture",
"document": "Entity: Cloud-Native Applications (CNA). Class: Application Architecture. characteristic: Highly Scalable..."
}
…and:
{
"entity_id": 209,
"class": "Application Architecture",
"document": "Entity: Microservices Architecture. Class: Application Architecture. characteristic: Modular Approach..."
}
Including both the entity name and class in the serialized text tends to improve semantic grouping and downstream filtering when performing hybrid vector+metadata queries.
Generating Vector Embeddings with Gemini
We’ll embed each synthesized document using Google’s gemini-embedding-001 model. Gemini supports configurable output dimensionality; here we choose 768 dimensions to balance semantic fidelity with storage footprint and ANN search cost. You can scale up or down depending on retrieval precision needs and budget.
Throughput Note: Free-tier rate limits apply; batching + polite sleeps help avoid throttling. Adjust batch_size and time.sleep() to match your quota.
GEMINI_MODEL_NAME = "gemini-embedding-001"
EMBEDDING_DIMENSIONS = 768
gemini_client = genai.Client(api_key=google_api_key)
def generate_embeddings_with_gemini(docs,
batch_size=96,
model_name=GEMINI_MODEL_NAME,
output_dim=EMBEDDING_DIMENSIONS,
rpm_delay=60):
"""Add 'embedding' (list[float]) to each doc in-place."""
texts = [d["document"] for d in docs]
for start in range(0, len(texts), batch_size):
batch_texts = texts[start:start+batch_size]
embed_cfg = types.EmbedContentConfig(
output_dimensionality=output_dim,
task_type="RETRIEVAL_DOCUMENT", # Recommended for corpus items
)
result = gemini_client.models.embed_content(
model=model_name,
contents=batch_texts,
config=embed_cfg,
)
batch_embs = [e.values for e in result.embeddings]
if len(batch_embs) != len(batch_texts):
print("Embedding count mismatch; skipping batch.")
continue
for j, emb in enumerate(batch_embs):
docs[start + j]["embedding"] = emb
# simple rate-limit guard
time.sleep(rpm_delay)
docs_with_embs = [d for d in docs if "embedding" in d]
print(f"Generated embeddings for {len(docs_with_embs)} docs.")
return docs_with_embs
Run:
docs_with_embeddings = generate_embeddings_with_gemini(docs_to_embed)
print(len(docs_with_embeddings), "embedded docs")
print("Dim:", len(docs_with_embeddings[0]["embedding"]))
print("Head:", docs_with_embeddings[0]["embedding"][:5])
Example output:
Generated embeddings for 276 docs.
276 embedded docs
Dim: 768
Head: [-0.00747136 0.01111416 -0.00263643 -0.0615893 -0.00403898]
At this point we have an in-memory list of dicts: each entry carries (entity_id, class, document, embedding[]). Next we’ll create an ApertureDB DescriptorSet configured for 768-dim vectors (Cosine, HNSW), ingest the vectors in parallel, and link each descriptor back to its source entity, unlocking fast semantic recall with graph-native joins.
Creating the Vector Index (DescriptorSet)
ApertureDB stores vectors inside DescriptorSets.
When you create a set, you must lock in the dimension. The distance metric and search engine can be left flexible, allowing multiple options if needed.
For GraphRAG, the dimensions are set to 768 to match the output from Gemini. The metric used is cosine similarity (CS), which is well-suited for dense semantic embeddings. The engine chosen is HNSW because it provides fast and memory-efficient approximate nearest neighbor (ANN) search.
ApertureDB lets you mix-and-match metrics/engines and even store several per set, but a single HNSW-cosine index is plenty for our use-case. (ApertureDB Documentation)
DESCRIPTOR_SET_NAME = "entity_embeddings_gemini"
def create_vector_index(client, set_name, dims):
"""Create a DescriptorSet (vector index)."""
add = [{
"AddDescriptorSet": {
"name": set_name,
"dimensions": dims,
"metric": "CS", # cosine
"engine": "HNSW" # hierarchical NSW
}
}]
resp, _ = client.query(add)
assert resp[0]["AddDescriptorSet"]["status"] == 0
print(f"Created DescriptorSet '{set_name}'.")
create_vector_index(client, DESCRIPTOR_SET_NAME, EMBEDDING_DIMENSIONS)
Ingesting Embeddings at Scale
The ParallelLoader helper takes an iterator of (query, blobs) tuples, batches them, and writes to the database with a configurable thread-pool. This drives ingestion throughput into the hundreds of descriptors per second on modest hardware.
def ingest_embeddings(client, set_name, docs):
"""Convert each embedding to bytes and upload as Descriptor."""
data = []
for d in docs:
q = [{
"AddDescriptor": {
"set": set_name,
"properties": {
# Store source metadata *on* the descriptor
"source_entity_id": d["entity_id"],
"source_entity_class": d["class"],
}
}
}]
blob = [np.array(d["embedding"], dtype=np.float32).tobytes()]
data.append((q, blob))
loader = ParallelLoader(client)
loader.ingest(generator=data, batchsize=64, numthreads=8, stats=True)
ingest_embeddings(client, DESCRIPTOR_SET_NAME, docs_with_embeddings)
Each element corresponds to one AddDescriptor call; we now have 276 vectors sitting in entity_embeddings_gemini.
Linking Descriptors Back to the Graph (has_embedding edges)
Vector search alone is not GraphRAG.
We need an explicit edge that lets us jump from a semantic hit → the originating entity → that entity’s neighbors. The pattern we’ll follow is: (Entity)-[:has_embedding]->(Descriptor)
def connect_embeddings(client, set_name, docs):
"""Create (Entity)<-[:has_embedding]-(Descriptor) edges."""
queries = []
for d in docs:
eid, eclass = d["entity_id"], d["class"]
q = [
{ # find the entity node
"FindEntity": {
"_ref": 1,
"with_class": eclass,
"constraints": {"id": ["==", eid]},
}
},
{ # find its descriptor (by metadata)
"FindDescriptor": {
"_ref": 2,
"set": set_name,
"constraints": {"source_entity_id": ["==", eid]},
}
},
{ # connect them
"AddConnection": {
"class": "has_embedding",
"src": 1,
"dst": 2
}
}
]
queries.append((q, []))
loader = ParallelLoader(client)
loader.ingest(generator=queries, batchsize=64, numthreads=8, stats=True)
connect_embeddings(client, DESCRIPTOR_SET_NAME, docs_with_embeddings)
Loader output confirms 276 AddConnection commands succeeded - the same number as our documents.
What We Achieved
- 768-dim Gemini embeddings for every entity (≈ 276 vectors).
- HNSW-cosine DescriptorSet in ApertureDB.
- Batch ingestion using ParallelLoader.
- has_embedding edges linking each descriptor to its source entity.
With these pieces in place we can issue hybrid queries:
- Vector similarity → returns a descriptor id.
- Traverse has_embedding-in → fetch the entity.
- Graph hops → pull in first-ring neighbors (properties, relationships, provenance).
This enables Graph-aware RAG: answers are seeded by dense semantic recall and enriched with structured graph context, improving grounding and interpretability.
Next Up - Part 4: End-to-End GraphRAG Retrieval & Performance Validation
In the next post we will:
- Write a small retriever that:
- Accepts a natural-language question.
- Calls the embedding model (embed_query) to vectorize it.
- Performs an ANN search on entity_embeddings_gemini.
- Accepts a natural-language question.
Expands results via graph traversal (FindConnection, FindEntity), and validate the performance of GraphRAG against vector RAG.
Stay tuned!
References
- Colab Notebooks: Cloud Version and Local (udocker) Version
- Github Repo with all Notebooks
- Google’s Gemini Docs
- ApertureDB Docs
Blog Series
- Automating Knowledge Graph Creation with Gemini and ApertureDB - Part 1
- Automating Knowledge Graph Creation with Gemini and ApertureDB - Part 2
Ayesha Imran | LinkedIn | Github
I am a software engineer passionate about AI/ML, Generative AI, and secure full-stack app development. I’m experienced in RAG and Agentic AI systems, LLMOps, full-stack development, cloud computing, and deploying scalable AI solutions. My ambition lies in building and contributing to innovative projects with real-world impact. Endlessly learning, perpetually building.
Images by Volodymyr Shostakovych, Senior Grahphic Designer













.png)
.png)















.jpeg)





.png)


