Technologies like RAG (retrieval-augmented generation), semantic search systems, and generative applications wouldn’t be possible without vector databases. A very few of these databases, such as ApertureDB, are truly capable of natively handling more than just text. They now work with images, audio, and other data types, which opens up new possibilities across industries like healthcare, retail, and finance.
For building this example, we pick healthcare advertising because it shows a great blend of multimodality. With strict rules around accuracy, disclosure, and patient privacy, it’s critical to include all Material Facts in marketing content. These are details that could influence a patient’s understanding or choices.
In this blog, we will discuss how a combination of ApertureDB, Unstructured, and OpenAI can help detect and flag missing material facts in healthcare advertisements.
The Landscape of Multimodality and Vector Databases
Data needed for AI is no longer just rows and columns in a structured database. Today's applications require multimodal data - medical records with scanned documents, handwritten notes, diagnostic images, and structured lab results all in one system.
While companies traditionally used polyglot persistence (MySQL for transactions, Elasticsearch for search, Redis for caching), they are now seeking multimodal databases that natively support multiple data types in one unified location.
Vector databases form a key component of such databases and have become essential for AI-driven applications that need semantic search across unstructured data. Rather than exact matches, they enable similarity-based retrieval through embeddings¹.
Key advantages:
- Fast similarity search based on meaning, not keywords
- Horizontal scalability for large AI applications
- Multimodal data support
- Cost-optimized serverless architectures
¹ For a deeper dive into multimodal data handling and vector database fundamentals, see our earlier posts.
This cuts it down significantly while maintaining the core message. The reference to your earlier content lets readers dive deeper if needed without rehashing basics here.
The Healthcare Sector and the Impact of AI
Within the healthcare sector, healthcare marketing faces intense scrutiny, especially around prescription drug advertising and could benefit the most from AI. The FDA demands accurate claims and prohibits off-label promotion. Any violations can be costly. GlaxoSmithKline paid $3 billion in 2012, while Pfizer faced a $2.3 billion settlement in 2009 for misleading marketing practices.
In addition to federal regulations, state laws often introduce more requirements. Some states need special licenses for advertising healthcare services. This makes compliance even more complex. To manage these risks, healthcare organizations need strong compliance programs, ongoing staff training, and legal support.
AI is already making a significant impact across the healthcare sector. It helps doctors detect diseases earlier, tailor treatments to individual patients, and improve efficiency in scheduling and billing. It also plays an important role in compliance. AI can review large volumes of regulations, identify missing information, and help ensure that communications meet legal standards. This is especially useful in healthcare marketing, where accuracy and transparency are essential.
In the next section, we will discuss how tools like ApertureDB, Unstructured, and OpenAI can help verify that healthcare advertisements include all required material facts.
Problem Statement: Avoiding Omission of Material Facts in Healthcare Advertising with AI
A single social media post can influence millions. This is exactly what happened when Kim Kardashian promoted Diclegis, a morning sickness prescription drug. She shared her positive experience, calling it safe and effective for her followers. However, the problem with her post was that she omitted a crucial fact: Diclegis had never been studied in women with hyperemesis gravidarum, a severe form of morning sickness.


The FDA issued a warning letter pointing out the missing fact. This example highlights a larger issue. How can we ensure that healthcare ads remain compliant?
Brands generate substantial amounts of content across various channels, including influencer partnerships. But unlike traditional ads, these posts often skip formal reviews, which makes it easier to miss critical facts, with potentially harmful consequences.
Implementation
To address material fact omissions, we developed a two-part solution:
- Identifying Omissions: We prompt an LLM to detect potential omissions. The model identifies missing details, including known limitations, contraindications, and required evidence. The prompt is designed to reduce the risk of hallucinated outputs.
- Cross-Referencing: Using the ApertureDB vector database, we perform similarity searches against clinical PDFs with medical-optimized embeddings for accurate validation.
Architecture
Our approach has two key parts: Ingestion and Cross-referencing.
Ingestion: Storing Medical And Clinical Information in a Multimodal AI Database

We pull text and images from clinical PDFs, split the content into smaller pieces, embed them, and store everything in ApertureDB. Since these are medical documents, we use a fine-tuned embedding model optimized for medical benchmarks. This ensures more accurate and relevant results.
Cross-Referencing: Verifying Omissions with Trusted Sources
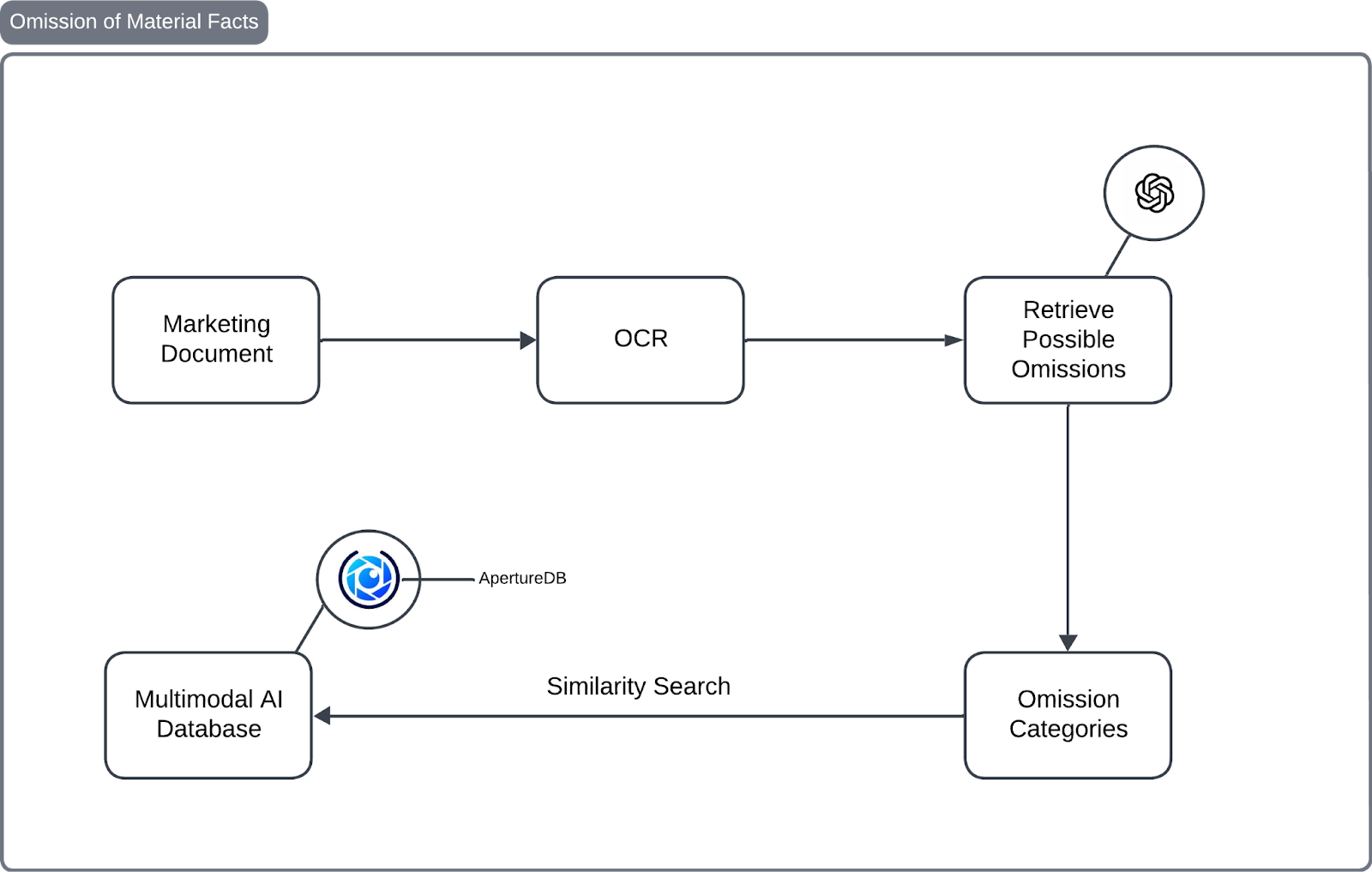
After an LLM flags potential omissions, we verify them against trusted clinical PDFs. Using ApertureDB, we run similarity searches to cross-check the facts and confirm their accuracy. This step ensures that the final content is both complete and reliable. Now, let's walk through the process of implementing it.
Why ApertureDB Fits Our Needs
In building our system for clinical documents, images, and tables, we needed more than vector search. We needed a database that stores source data, maintains rich relationships, and lets us query across both vectors and metadata. ApertureDB delivers this.
- Multimodal storage: Images, text, tables, embeddings, and metadata live together. You can retrieve an embedding and get its source instantly.
- Graph-backed relations: Pages, chunks, tables, and images are inherently linked. We can query and filter through these connections. With ApertureDB’s in-memory property graph, we can represent these relations and query them (filter, traverse) as part of our retrieval logic.
- Unified querying: We can combine similarity search, metadata filters, and graph constraints in one step. This allows for specific searches, such as “only images from page X” or “only chunks from pages with certain metadata”.
- Traceability: Everything is stored with context, making it easy to audit, debug, and visualize results.
Let’s see it in action.
Setup
Setting up the environment correctly is crucial to ensure the smooth execution of the project. This section will guide you through installing dependencies, understanding the folder structure, and configuring essential settings.
Install Dependencies
The project relies on various libraries for optical character recognition (OCR) (ApertureDB AI Workflows now automate OCR on PDFs but it was launched after we created these examples), natural language processing (NLP), and document processing. These dependencies are listed in requirements.txt. To install them, run:
pip install -r requirements.txtRequired Packages
Here's a breakdown of the key dependencies:
easyocr– Optical character recognition (OCR) for extracting text from images.pyyaml– Parsing YAML configuration files.textblob– Basic NLP processing.python-dotenv– Managing environment variables securely.unstructured[pdf]– Extracting text from PDFs.aperturedb– Multimodal database for similarity search and document management.sentence_transformers– Creating embeddings for semantic similarity.PyMuPDF / fitz– Handling and parsing PDF files.
Configure the Project
The main configuration file is located at config/config.yaml and contains:
embedding_model: "clip-ViT-B-32"
collection_name: "omission_db_v2"
marketing_claim_image: "data/KimKardashianAd.png"
reference_clinical_pdf: "data/DiclegisClinicalInformation.pdf"ApertureDB now supports tokens which can replace the user and password combination. Read more about the environment setup in their documentation.
Key Configuration Elements
- marketing_claim_image & reference_clinical_pdf – Paths to documents used for comparison.
- embedding_model – Specifies the embedding model used for text similarity and image similarity.
- collection_name – The name of the vector database collection.
Ensure that all necessary credentials are up to date before running the project. Now, let’s write some preprocessing logic.
Preprocessing
The first page of the clinical document for Diclegis includes key medical details that were missing from the promotional content. Some of the most important sections are:

Contraindications– Lists conditions under which Diclegis should not be used.Use in Specific Populations– Details how the drug affects different demographic groups, including pregnant women and elderly patients.Warnings and Precautions– Highlights safety concerns and possible risks.Adverse Reactions– Provides information on potential side effects.Dosage and Administration– Guides correct drug usage.
We will use the Unstructured library to extract the important sections and pages from the above information for cross-referencing.
Preprocessing PDFs
To efficiently preprocess the PDF and extract relevant information, including tables and text, we use the following code:
#preprocessor/extract.py
from unstructured.partition.pdf import partition_pdf
import easyocr
import os
class Processor():
def __init__(self) -> None:
self.model = easyocr.Reader(['en']) # Load model
def clean_text(self, ocr_output):
"""
Extracts text from OCR output and applies spell correction.
Args:
ocr_output (list): The OCR output containing bounding boxes, text, and probabilities.
Returns:
list: A list of corrected text strings.
"""
corrected_texts = []
for item in ocr_output:
if len(item) > 1: # Ensure the item has text content
text = item[1] # Extract the text
corrected_texts.append(text)
return " ".join(corrected_texts)
def extract(self, document):
"""
Extracts text from a document based on its type.
Args:
document (str): Path to the document file.
Returns:
list: Extracted text if the document is an image; None for unsupported types.
"""
if not os.path.exists(document):
raise FileNotFoundError(f"The document '{document}' does not exist.")
file_extension = os.path.splitext(document)[1].lower()
if file_extension in ['.png', '.jpg', '.jpeg']: # Supported image formats
result = self.model.readtext(document)
return result
elif file_extension == '.pdf':
result = partition_pdf(document, infer_table_structure=True, strategy='hi_res', languages=["eng"])
return result
else:
print(f"Unsupported file type: {file_extension}")
return NoneThis Processor class extracts text from PDFs and images using EasyOCR for images and unstructured.partition.pdf for PDFs. It also includes a method to clean OCR output by extracting and joining the recognized text.
Extracting Images from PDF
The following utility function extracts images from the PDF, allowing further analysis:
#extras/utils.py
def extract_images(pdf_path, output_folder="extracted_images"):
doc = fitz.open(pdf_path)
os.makedirs(output_folder, exist_ok=True)
images_info = []
for page_num in range(len(doc)):
for img_index, img in enumerate(doc[page_num].get_images(full=True), start=1):
xref = img[0] # XREF index
base_image = doc.extract_image(xref)
image_bytes = base_image["image"]
image_ext = base_image["ext"]
image_filename = f"{output_folder}/page_{page_num+1}_img_{img_index}.{image_ext}"
with open(image_filename, "wb") as img_file:
img_file.write(image_bytes)
images_info.append({"page": page_num+1, "image": image_filename})
return images_infoNote: We will use the functions above to also extract information from the marketing document.
Building the Vector Indexes
The VectorStore class is responsible for managing embeddings and images within ApertureDB. This section breaks down the functionality of each method in the class.
Initializing the VectorStore
#storage/db.py
class VectorStore:
def __init__(self, host: str, user: str, password: str, collection_name: str):
"""
Initializes the ApertureDB client.
:param host: The database instance name or IP (without http://).
:param user: Username for authentication.
:param password: Password for authentication.
"""
self.client = create_connector(key = os.getenv("APERTUREDB_API_KEY"))
self.client.query([{"GetStatus": {}}]) # Verify connection
self.descriptorset_name = collection_nameThis constructor establishes a connection with ApertureDB using the provided credentials. It also verifies that the connection is successful by sending a status check query.
Setting Up a Collection
def set_collection(self, collection_name: str, dimensions: int = 1024):
"""
Sets the descriptor set (collection) to be used. If it doesn't exist, it creates one.
:param collection_name: Name of the descriptor set.
:param dimensions: Dimensionality of the embeddings.
"""
self.descriptorset_name = collection_name
q = [{
"AddDescriptorSet": {
"name": self.descriptorset_name,
"dimensions": dimensions,
"engine": "HNSW",
"metric": "CS",
"properties": {
"year_created": 2025,
"source": "ApertureDB dataset",
}
}
}]
return self.client.query(q)This method defines a descriptor set (vector collection) and initializes it if it doesn’t already exist. It allows specifying dimensionality and metadata.
Ingesting Embeddings
def ingest_embeddings(self, embeddings: np.ndarray, ids: list, metadatas: list = None):
"""
Ingests embeddings along with metadata into ApertureDB.
:param embeddings: The embeddings (as a NumPy array) to be stored.
:param ids: A list of unique IDs for each embedding.
:param metadatas: A list of metadata dictionaries for each embedding.
"""
if self.descriptorset_name is None:
raise ValueError("Descriptor set is not set. Use 'set_collection' first.")
queries = []
blobs = []
for idx, embedding in enumerate(embeddings):
embedding = np.array(embedding, dtype=np.float32).tobytes()
metadata = metadatas[idx] if metadatas else {}
q = {
"AddDescriptor": {
"set": self.descriptorset_name,
"label": metadata.get("_label", "unknown"),
"properties": {"id": ids[idx],
**metadata},
"if_not_found": {"id": ["==", ids[idx]]}
}
}
queries.append(q)
blobs.append(embedding)
return self.client.query(queries, blobs)This method ingests embeddings into the vector database, along with metadata and unique IDs, for easy retrieval. We also ingest image embeddings into the vector database for the clinical document, including crucial visual data such as efficacy charts, safety profiles, and statistical graphs that traditional text analysis may miss.
Querying Similar Embeddings
def query_embeddings(self, query_embedding: np.ndarray, top_k: int = 5, return_images: bool = True):
"""
Queries the ApertureDB for similar embeddings.
If results are images and `return_images=True`, also fetch the image blobs.
:param query_embedding: The query embedding to search for similar items.
:param top_k: Number of similar embeddings to return.
:param return_images: Whether to fetch actual images for image descriptors.
:return: List of results with id, label, metadata, and optional image blob.
"""
if self.descriptorset_name is None:
raise ValueError("Descriptor set is not set. Use 'set_collection' first.")
embedding_bytes = query_embedding.astype("float32").tobytes()
q = [{
"FindDescriptor": {
"set": self.descriptorset_name,
"k": top_k,
"return": ["id", "label", "properties"]
}
}]
responses, _ = self.client.query(q, [embedding_bytes])
descriptors = responses[0]["FindDescriptor"]["descriptors"]
results = []
for d in descriptors:
result = {
"id": d["properties"]["id"],
"label": d["label"],
"metadata": d["properties"]
}
# If this is an image descriptor and we want blobs, fetch the image
if d["label"] == "image" and return_images:
q_img = [{
"FindImage": {
"constraints": {"id": ["==", d["properties"]["id"]]},
"blobs": True,
"results": {"limit": 1}
}
}]
resp, blobs = self.client.query(q_img)
if blobs:
result["image_blob"] = blobs[0]
results.append(result)
return resultsThis function searches ApertureDB for items with embeddings similar to a given query embedding. It converts the query into bytes, performs a nearest-neighbor search in the specified descriptor set, and retrieves the top k matching descriptors along with their metadata. If the results are images and return_images is True, it also fetches the actual image data for each match.
Another way to simplify the query is to connect images with their embeddings and follow the graph connections.
Ingesting Data into the Vector Store
The ingestion script (storage/ingest.py) processes and stores document elements as embeddings in the vector store.
- Initialize
VectorStorewith credentials - Extract text and images from a PDF
- Chunk document by title
- Process tables
- Associate extracted data with page numbers
- Store embeddings and images
config = read_yaml(CONFIG_PATH)
vector_store = VectorStore(
collection_name=config.get("collection_name"),
)
vector_store.set_collection(dimensions=512)
pdf_path = config.get("clinical_doc")
images_info = extract_images(pdf_path=pdf_path)
processor = Processor()
clinical_doc_elements = processor.extract(document=pdf_path)
chunks = chunk_by_title(clinical_doc_elements)
tables = [el for el in clinical_doc_elements if el.category == "Table"]
page_data = {}
for chunk in chunks:
page_number = chunk.metadata.page_number
if page_number not in page_data:
page_data[page_number] = {"text": [], "table": None, "image": None}
page_data[page_number]["text"].append(chunk.text)
for table in tables:
page_number = table.metadata.page_number
if page_number in page_data:
page_data[page_number]["table"] = table
for idx, image_info in enumerate(images_info, start=1):
page_number = image_info["page"]
if page_number in page_data:
page_data[page_number]["image"] = {
"filename": image_info["image"],
"id": f"img_{idx}",
"page": page_number
}
embeddings, ids, metadatas = [], [], []
for page_number, data in page_data.items():
combined_text = "\n".join(data["text"])
if combined_text.strip():
emb = get_multimodal_embedding(combined_text, is_image=False)
embeddings.append(emb)
ids.append(f"text_page_{page_number}")
metadatas.append({
"type": "text",
"page_number": page_number,
"text": combined_text
})
if data["table"]:
table_text = data["table"].text
emb = get_multimodal_embedding(table_text, is_image=False)
embeddings.append(emb)
ids.append(f"table_page_{page_number}")
metadatas.append({
"type": "table",
"page_number": page_number,
"table": table_text
})
if data["image"]:
emb = get_multimodal_embedding(data["image"]["filename"], is_image=True)
embeddings.append(emb)
ids.append(data["image"]["id"])
metadatas.append({
"type": "image",
"page_number": page_number,
"image": data["image"]["filename"]
})
vector_store.ingest_embeddings(embeddings, ids, metadatas)This script structures document elements into a retrievable vector format, making them searchable within ApertureDB. Before we proceed to identify and extract the omissions, it’s a good time to discuss the embedding model used.
For the image embedding model, we use CLIP ViT-B/32, an open-source vision-language model that maps images and text into a shared 512-dimensional space. It achieves around 63% zero-shot accuracy on ImageNet, enabling strong performance in cross-modal retrieval and classification tasks. We have written an inference function for generating multimodal embeddings:
from sentence_transformers import SentenceTransformer
from extras.utils import read_yaml
from extras.constants import CONFIG_PATH
import numpy as np
from PIL import Image
config = read_yaml(CONFIG_PATH)
model = SentenceTransformer(config.get("multimodal_embedding_model"))
def get_multimodal_embedding(input_data, is_image=False):
if is_image:
img = Image.open(input_data)
return model.encode(img, convert_to_tensor=True)
else:
return model.encode(input_data, convert_to_tensor=True)Let’s move on to the most important discussion of logic, detecting omissions in marketing documents.
Detecting Omissions in Medical Marketing Documents
This section details how omissions are identified using an LLM, cross-referenced with clinical data, and validated for accuracy. The first step in detecting omissions is extracting potential gaps in the medical marketing document. This is accomplished using an LLM that analyzes the text against predefined omission categories.
Defining Omission Categories
The categories of omissions are structured using a Pydantic model:
from pydantic import BaseModel
from typing import List
class MedicalOmissionInfo(BaseModel):
omitted_side_effects_and_risks: List[str]
omitted_contraindications: List[str]
omitted_safety_information: List[str]
omitted_efficacy_and_limitations: List[str]
omitted_clinical_evidence: List[str]This structure ensures that all possible omission types are categorized systematically.
Extracting Omissions with an LLM
The OmissionExtractor class uses OpenAI's LLM to identify missing information based on the above categories:
from openai import OpenAI
from dotenv import load_dotenv
from omission.models import MedicalOmissionInfo
load_dotenv()
client = OpenAI()
class OmissionExtractor:
def __init__(self, model: str = "gpt-4o-2024-08-06"):
self.model = model
def extract(self, text: str) -> MedicalOmissionInfo:
prompt = (
"Analyze the document and assess whether critical information is omitted under the following categories. "
"Provide a general statement for each category about whether omissions are present and what their potential effect might be:"
"\n- Side Effects and Risks"
"\n- Contraindications"
"\n- Safety Information"
"\n- Efficacy and Limitations"
"\n- Clinical Evidence"
)
completion = client.beta.chat.completions.parse(
model=self.model,
messages=[
{"role": "system", "content": prompt},
{"role": "user", "content": text},
],
response_format=MedicalOmissionInfo,
)
return completion.choices[0].message.parsedThe extractor processes a marketing document and returns structured information on potential omissions.
Cross-Referencing Omissions with Clinical Data
Once omissions are identified against the predefined categories, we ensure that the corresponding information is present in the clinical document. If yes, we flag it as an omission.
Checking for Omission Consistency
The MedicalOmissionChecker verifies whether omissions are supported by clinical evidence.
from typing import List, Dict, Tuple
from extras.constants import CONFIG_PATH
from extras.utils import read_yaml
from pydantic import BaseModel
from storage.db import VectorStore
from embedder.multimodal_embedding import get_multimodal_embedding
from omission.models import MedicalOmissionInfo
from colorama import Fore, Style
from dotenv import load_dotenv
from openai import OpenAI
from pydantic import BaseModel
from typing import Literal
load_dotenv()
class ConsistencyCheck(BaseModel):
status: Literal["Omission", "Fine", "No documents found"]
reason: str
class MedicalOmissionChecker:
def __init__(self, collection_name: str):
config = read_yaml(CONFIG_PATH)
self.vector_store = VectorStore(collection_name)
self.vector_store.set_collection()
self.client = OpenAI()
def _query_observation(self, observations: List[str]) -> Dict[str, List[str]]:
"""Query Aperture DB for each claim and return relevant documents."""
relevant_docs = {}
for observation in observations:
embeddings = get_multimodal_embedding(observation)
documents = self.vector_store.query_embeddings(embeddings)
relevant_docs[observation] = documents
return relevant_docs
def _check_consistency(self, post: str, observation: str, category: str, documents: list[str]) -> ConsistencyCheck:
"""Use an LLM to determine if the observation and documents are consistent."""
if not documents:
return ConsistencyCheck(status="No documents found", reason="No supporting references available")
prompt = f"""
This is the post: {post}
Your task is to evaluate whether the post omits important information.
Observation category: {category}
Observation: {observation}
Supporting documents:
{documents}
Decide if the documents support the observation or not:
- If yes, return status "Omission"
- If not, return status "Fine"
Provide also a short explanation in plain text.
"""
response = self.client.responses.parse(
model="gpt-4o-2024-08-06",
input=[
{"role": "system", "content": "You are a Medical Legal Reviewer. Output strictly as JSON."},
{"role": "user", "content": prompt},
],
text_format=ConsistencyCheck,
)
return response.output_parsed
def process_observation(self, post:str, observation_info: MedicalOmissionInfo) -> Dict[str, List[Tuple[str, str]]]:
"""Process all observation, cross-reference with Aperture DB, and check consistency."""
results = {}
observation_categories = {
"omitted_side_effects_and_risks": observation_info.omitted_side_effects_and_risks,
"omitted_contraindications": observation_info.omitted_contraindications,
"omitted_safety_information": observation_info.omitted_safety_information,
"omitted_efficacy_and_limitations": observation_info.omitted_efficacy_and_limitations,
"omitted_clinical_evidence": observation_info.omitted_clinical_evidence,
}
for category, observations in observation_categories.items():
if not observations:
results[category] = [("No observation provided", "No documents found")]
continue
relevant_docs = self._query_observation(observations)
category_results = []
for observation, documents in relevant_docs.items():
consistency = self._check_consistency(post, observation, category, documents)
category_results.append((observation, consistency))
results[category] = category_results
return results
def display_results(self, results: Dict[str, List[Tuple[str, "ConsistencyCheck"]]]):
"""
Display flagged claims in the terminal with appropriate colors.
Expects results in the form:
"""
for category, observations in results.items():
print(f"\nCategory: {category}")
for observation, result in observations:
status = result.status
reason = result.reason
if status == "Omission":
print(
Fore.RED
+ f"Observation: {observation}\n"
f" → Status: {status}\n"
f" → Reason: {reason}"
+ Style.RESET_ALL
)
elif status == "Fine":
print(
Fore.GREEN
+ f"Observation: {observation}\n"
f" → Status: {status}\n"
f" → Reason: {reason}"
+ Style.RESET_ALL
)
else:
print(
Fore.YELLOW
+ f"Observation: {observation}\n"
f" → Status: {status}\n"
f" → Reason: {reason}"
+ Style.RESET_ALL
)Running the Full Detection Pipeline
The following script integrates the extraction and validation steps:
from extras.utils import read_yaml
from preprocessor.extract import Processor
from omission.extract_omission import OmissionExtractor
from omission.check_omission import MedicalOmissionChecker
if __name__ == "__main__":
config = read_yaml(CONFIG_PATH)
processor = Processor()
checker = MedicalOmissionChecker(collection_name=config.get("collection_name"))
omission_extractor = OmissionExtractor()
marketing_post_text = processor.extract(config.get("marketing_claim_image"))
marketing_post_text_cleaned = processor.clean_text(marketing_post_text)
observation_info = omission_extractor.extract(marketing_post_text_cleaned)
results = checker.process_observation(marketing_post_text_cleaned, observation_info)
checker.display_results(results)The results are as follows:
Category: omitted contraindicationsObservation: There is no mention of specific health conditions or demographic populations for whom the drug might be dangerous or ineffective. → Status: Omission - The provided documents highlight important safety considerations and potential risks associated with Diclegis, such as somnolence, the need to avoid alcohol and other sedating medications, and the risk of overdose in children. These potential risks and specific conditions for use, such as avoiding activities requiring mental alertness and the pediatric use warning, are not mentioned in the post. Therefore, the omission of contraindications and safety information in the post is confirmed.
Category: …
Observation: …
We can see that our system identifies various omissions, including the one mentioned in the FDA's warning letter. Most of the technical part of the code is explained, but here is the GitHub repo for you. Simply place your documents in the configuration and run it.
Conclusion
Healthcare advertising relies on trust because accurate information can impact lives. Strict regulations make compliance essential. Multimodal data parsing and vector databases help by detecting missing details, verifying facts, and ensuring transparency without manual reviews. Automating this process saves time and builds credibility. Patients need reliable information, and AI helps ensure that they receive it.
In this guide, we used Unstructured and ApertureDB to parse PDFs and store embeddings, images, and text for better compliance monitoring. You can create a free ApertureDB trial account to explore its features.
As regulations grow stricter, multimodal AI tools are no longer optional. They are essential for the future of use cases such as healthcare marketing.














.png)
.png)















.jpeg)





.png)


